Genetic Algorithms
A genetic algorithm (GA) simulates evolution to try to find an optimum solution to a problem. It starts with a large random population of genomes. The actual format of the genome depends on the application. For example, if you were trying to create a sentence, the genome could be string, or, if you wanted to find rules to play the stock market, the genome could be an array of numbers representing possible rules. Once the initial population is created, the genomes are ranked using a fitness function. This function is a mathematical way of describing how successful the genome is at solving the problem. Next, a new generation of genomes is created by combining the more successful genomes. The theory is that if a genome is successful, it will statistically be used more frequently as a parent for the next generation, passing successful genes, or components of itself, to more solutions in the next generation. Thus, features that cause success will become slightly more abundant in each generation. Over many generations, successful features slowly emerge and combine to develop optimum solutions.
It is also possible to allow small amounts of mutation in each generation. This could be done by changing some random genes, adding new genes to a genome or removing existing genes from a genome. For example, if the genome were an array of numbers, then changing a gene would be changing the value of one of the numbers, adding a gene would be adding a number to the array and removing a gene would be removing a number from the array. Mutation stops the population from converging too quickly to a good, but not optimum solution. After many generations without mutation, a population could be so saturated with a few promising features, that other features cannot develop. Mutation ensures a certain degree of variety in each generation.
GAs have proven to be very robust for many applications. However, some conditions must be met for the algorithm to be successful. First, the problem must be encoded in such a way so that genomes can be easily combined and manipulated. A common way to do this is to have an array of numbers, then genomes can be combined using dual point crossover and mutated by changing/adding/removing numbers. Dual point crossover is done by choosing two parents and picking an index in each array to act as the crossover point. Then, one child is created by using the numbers from the first parent before its crossover point and the numbers from the second parent after its crossover point. A second child is created by using the two segments of the parent not already used.
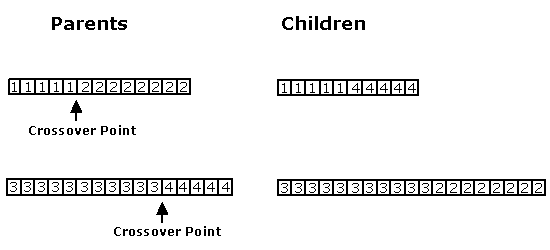
Second, the fitness function must return a large variety of values. If there are too few values, then it will be more difficult to jump to a higher value. In the extreme case of a boolean fitness function, the only way to find a solution is to arrive at it completely randomly as there is no way to build the solution in small steps. This completely defeats the purpose of a GA.