Autoencoders¶
A more complicated approach, but not always one that works better.
Idea: use deep learning to learn two nonlinear models, one of which (the encoder, $\phi$) goes from our original data in $\R^d$ to a compressed representation in $\R^r$ for $r < d$, and the other of which (the decoder, $\psi$) goes from the compressed representation in $\R^r$ back to $\R^d$.
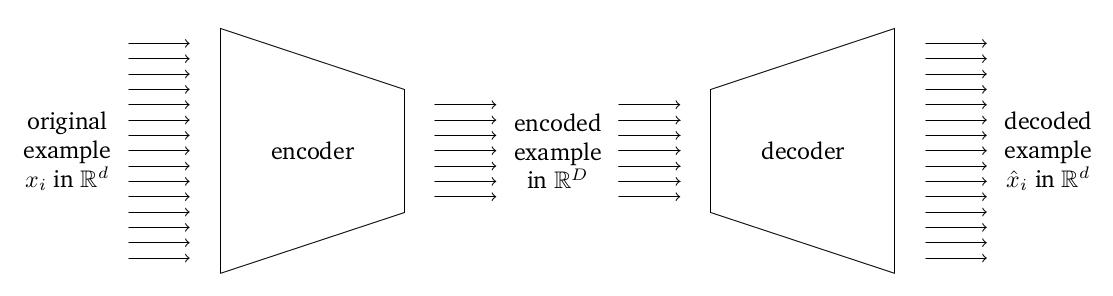
We want to train in such a way as to minimize the distance between the original examples in $\R^d$ and the "recovered" examples that result from encoding and then decoding the example. Formally, given some dataset $x_1, \ldots, x_n$, we want to minimize
$$\frac{1}{n} \sum_{i=1}^n \norm{ \psi(\phi(x_i)) - x_i }^2$$over some parameterized class of nonlinear transformations $\phi: \R^d \rightarrow \R^r$ and $\psi: \R^r \rightarrow \R^d$ defined by a neural network.